Higher-Order FCS Method Development | DNA & RNA Biophysics | Quantum Dot Photophysics | Super-Resolution SMLM Instrument Development
Higher-Order Fluorescence Correlation Spectroscopy (FCS) Method Development
FCS is an incredibly versatile technique convenient for probing the time-dependent properties of biological systems. In particular, FCS can be used to readout quantities such as concentration, translational diffusion coefficients, and hydrodynamic radii as well as provide information regarding the rotational dynamics and reaction kinetics within a system when an appropriate model is known.
However, routine FCS alone is limited in its capability to capture all pertinent reaction parameters associated with complex biochemical reactions, including two-state binding reactions or reactions in which the interacting components have different brightness. To overcome this limitation, our goal is to advance the higher-order FCS method that was originally proposed by Palmer and Thompson in the late 1980s and refers to the computation and analysis of third- or higher-order correlation functions in addition to the second-order functions obtained from routine analysis.
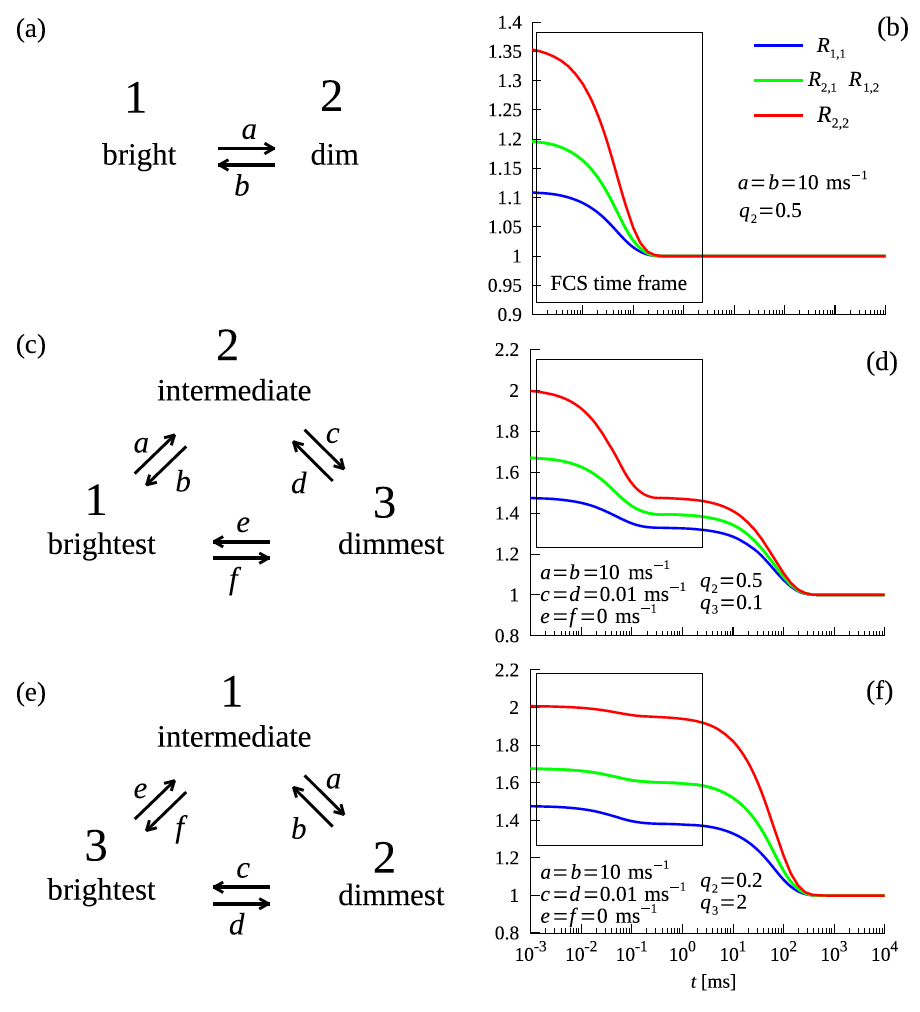
Example higher-order correlation functions with corresponding rate constants and brightness values describing the two- and three-state mechanism of DNA hairpin folding.
Recently, the group developed a sub-binning approach that provides a time resolution on the order of microseconds with a standard two-detector FCS setup and alleviates detector artifacts such as detector dead time and shot-noise, which are typically amplified in higher-order correlation analysis. As a proof-of-concept, the higher-order FCS method with sub-binning was applied to a model DNA hairpin system, from which a three-state folding mechanism.
DNA & RNA Biophysics
DNA and RNA molecules can adopt a variety of noncanonical conformations including hairpin structures. These hairpins are presumed to play an integral role in replication, transcription, and genetic recombination, though new information regarding their biological functions continues to be learned through kinetics and thermodynamics studies.
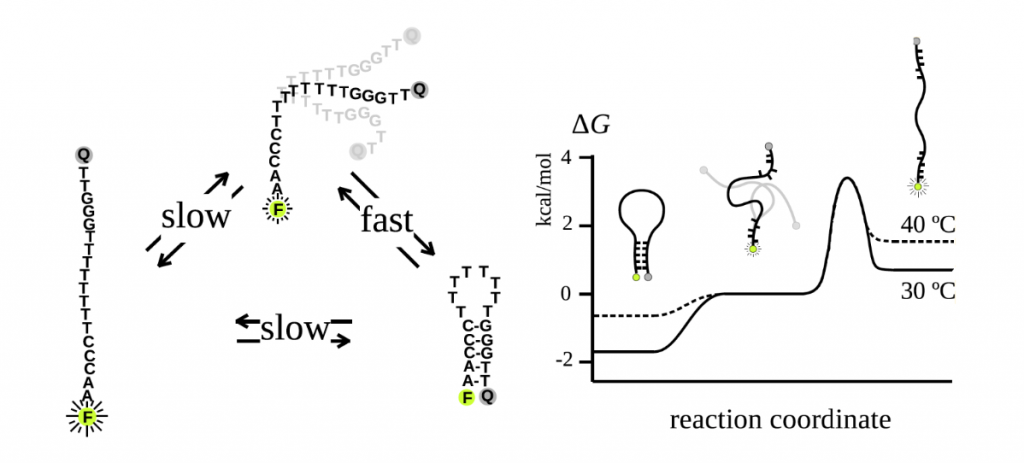
Our group is interested in investigating the full range of hairpin conformational states and probing the mechanism of hairpin folding, which was once thought to be a two-state mechanism but is now recognized as a three-state mechanism. Using higher-order FCS as well as dual-beam FCS in combination with stopped flow kinetic measurements, our work has not only validated a three-state folding model, but also demonstrated that external salt concentrations and temperature influence the folding kinetics and the free energy landscape.
To conduct FCS studies, single-stranded DNA is labeled with a fluorophore and fluorescence quencher, thereby enabling measurements of the fluorescence intensity fluctuations characteristic of the transitions between the folded, intermediate, and unfolded conformational states.
Quantum Dot Photophysics
Because quantum dots (QDs) offer desirable photophysical properties including high quantum yields, photostability, and tunable emission wavelengths, QDs are used in imaging, photodetectors, photovoltaics, and other applications. Our group has a particular interest in characterizing the electronic interactions among closely-spaced QDs using both wide-field and confocal optical microscopy techniques. We monitor photoluminescent properties such as emission intensity, polarization, color, lifetime, and other photon arrival statistics that paint a picture of the underlying dynamics. Most recently, we have probed energy transfer pathways within CdSe/CdS core/shell QD assemblies using time-resolved super resolution imaging and found evidence of energy donors and acceptors suggestive of a Förster resonance energy transfer mechanism.
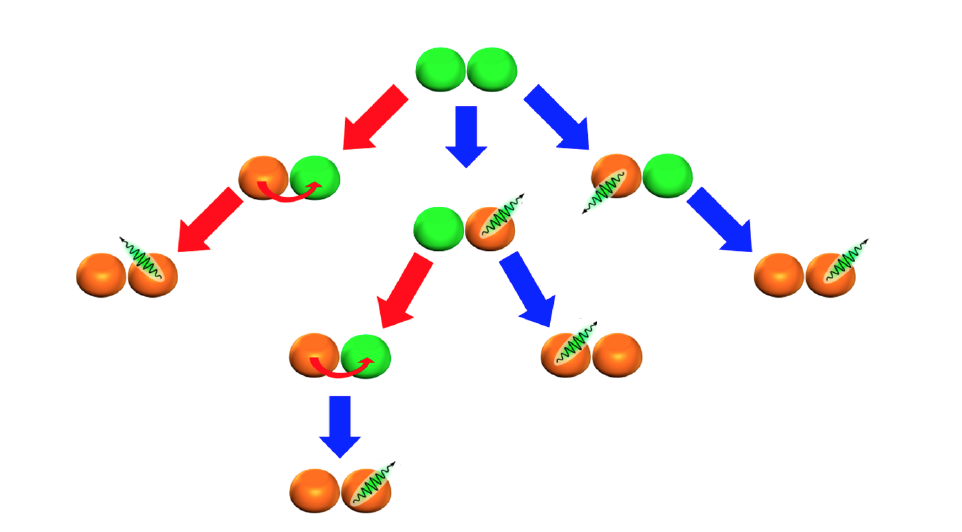
Pathways of photon emission (indicated by blue arrows) and energy transfer (indicated by red arrows) in a two-QD cluster in which green QDs have an exciton and orange QDs do not. Curved red arrows denote the direction of exciton transfer.
Super-Resolution Single-Molecule Localization Microscopy (SMLM) Instrument Development
Super-resolution imaging comprises a number of techniques that overcome the diffraction limit of a typical optical microscope in order to obtain high-resolution images. Within this toolkit of techniques, SMLM is one example.
SMLM typically relies upon a CCD or CMOS camera to collect the fluorescence or photoluminescence emission from a sample of blinking particles whose reversible transitions between an activated state and a dark state aid in resolving closely-packed multiple emitters within the sample.
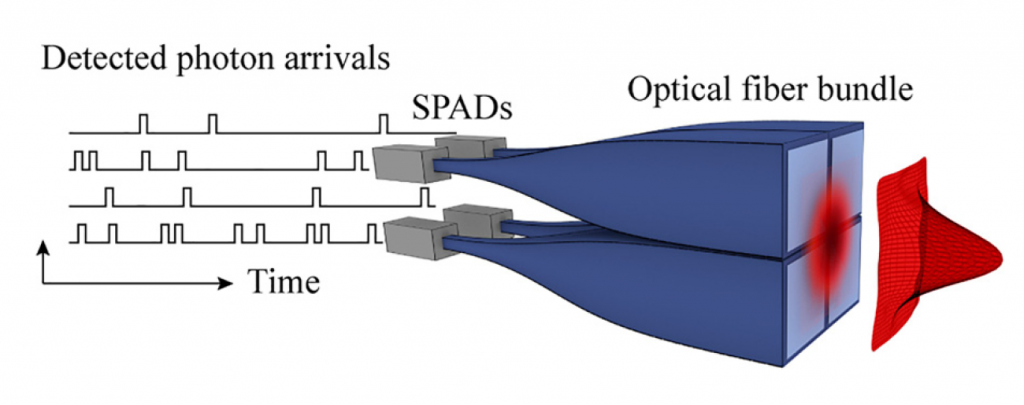
Our group has recently developed a detection scheme for a single-molecule localization microscope incorporating four single-photon avalanche photodiode (SPAD) detectors and a time-correlated single-photon counter (TCSPC), which was demonstrated by creating a spatial mapping of quantum dot positions based on emission intensity.